Developing new methods to manipulate fly genes offers a strong driving force to better understand their function. The main goal of our initiative is to develop a strategy that allows multiple manipulations of a fly gene in an integrated fashion using a single tool. The strategy that we developed utilizes CRISPR and is named CRIMIC (CRISPR-Mediated Integration Cassette). In simple terms, a CRIMIC insertion can allow us to

Some of these features are shown in Figure 1.
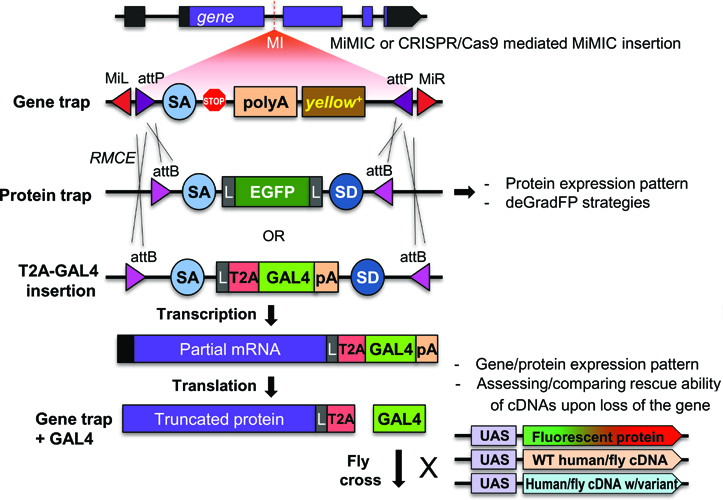
In the Gene Disruption Project, our lab creates the reagents needed to implement the above mentioned technologies for most genes. Currently,
more than 25,000 stocks created in our labs are distributed by the Bloomington Drosophila Stock Center (BDSC) worldwide. These stocks collectively allow manipulations of more than 70% of all fly genes. More than a million copies of these stocks have been distributed, proving the popularity and utility of our resources.
Over the past few years, we have tagged more than 3,000 genes with the multifunctional tags described above (see the Gene Disruption Project database). We are in the process of inserting these very versatile tags in thousands of additional genes. Our current strategy combines commercial DNA synthesis whit a highly efficient single step cloning to generate homology donor constructs that contain all the components required for successful CRISPR mediated homologous recombination. Over the years, we improved our targeting strategies to reach ~90% transgenic insertion efficiency. These insertions integrate a GAL4 gene into a gene of interest and these GAL4 insertions allow us, for example, to drive UAS-human cDNAs to assess if human cDNAs can rescue the fly loss of function mutation. To this end we are generating a collection of fly transgenic stocks that carry UAS-human cDNAs in collaboration with Sue Celniker (LBNL), Shinya Yamamoto (BCM) and Toshiyuki Takano-Shimizu (Kyoto Stock Center).
Kanca O, Zirin J, Hu Y, Tepe B, Dutta D, Lin WW, Ma L, Ge M, Zuo Z, Liu LP, Levis RW, Perrimon N, Bellen HJ (2022) An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR mediated homologous recombination. eLife 11:e76077. PMCID: PMC9239680.
Kanca O, Zirin J, Garcia-Marques J, Knight S, Yang-Zhou D, Amador G, Chung HL, Zuo Z, Ma L, He Y, Lin WW, Fang Y, Ge M, Yamamoto S, Schulze KL, Hu Y, Spradling AC, Mohr SE, Perrimon N, Bellen HJ (2019) An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. eLife 8:e51539. PMCID: PMC6855806.
Li-Kroeger D, Kanca O, Lee PT, Cowan S, Lee MT, Jaiswal M, Salazar JL, He Y, Zuo Z, Bellen HJ (2018) An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. eLife 7:e38709. PMCID: PMC6095692.
Lee PT, Zirin J, Kanca O, Lin W-W, Schulze KL, Li-Kroeger D, Tao R, Devereaux C, Hu Y, Chung V, Fang Y, He Y, Pan H, Ge M, Zuo Z, Housden BE, Mohr SE, Yamamoto S, Levis RW, Spradling AC, Perrimon N, Bellen HJ (2018) A gene-specific T2A-GAL4 library for Drosophila. eLife 7:e35574. PMCID: PMC5898912.
Nagarkar-Jaiswal S, Manivannan SN, Zuo Z, Bellen HJ (2017) A cell cycle-independent, conditional gene inactivation strategy for differentially tagging wild-type and mutant cells. eLife 6:e26420. PMCID: PMC5493436.
Nagarkar-Jaiswal S, DeLuca SZ, Lee PT, Lin W-W, Pan H, Zuo Z, Lv J, Spradling AC, Bellen HJ (2015) A genetic toolkit for tagging intronic MiMIC containing genes. eLife 4:e08469. PMCID: PMC4499919.
Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin W-W, He Y, Schulze KL, Booth BW, Evans-Holm M, Venken KJ, Levis RW, Spradling AC, Hoskins RA, Bellen HJ (2015) A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife 4:e05338. PMCID: PMC4379497.
Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, Bellen HJ (2011) MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature Methods 8:737-743. PMCID: PMC3191940. Covered in Nature Methods 8(9):728-9, doi: 10.1038/nmeth.1672, News & Views.
Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, Hoskins RA, Spradling AC (2011) The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188:731-743. PMCID: PMC3176542.